ACBO-Feat
AC BO Hackathon 2024
This is a repo created for the 2024 AC BO Hackathon
Project 8
BO for Drug Discovery: What is the role of molecular representation?
Project leads
- Fanjin Wang (University College London)@FrankWanger
Contributors
- Quinn Gallagher (Princeton University) @QGallagher
- Ankur Gupta (Lawrence Berkeley National Laboratory) @ankur56
- Christina Schenk (IMDEA Materials Institute) @schenkch
Questions we are asking (and answering):
- How fingerprints play a role in BO process
- Will dimensionality reduction help with BO performance?
- Instead of GP surrogate, will I get better results with a random forest surrogate?
Video
See our video presentation. Comment, suggest, and vote project 8 in the judging session!
Poster
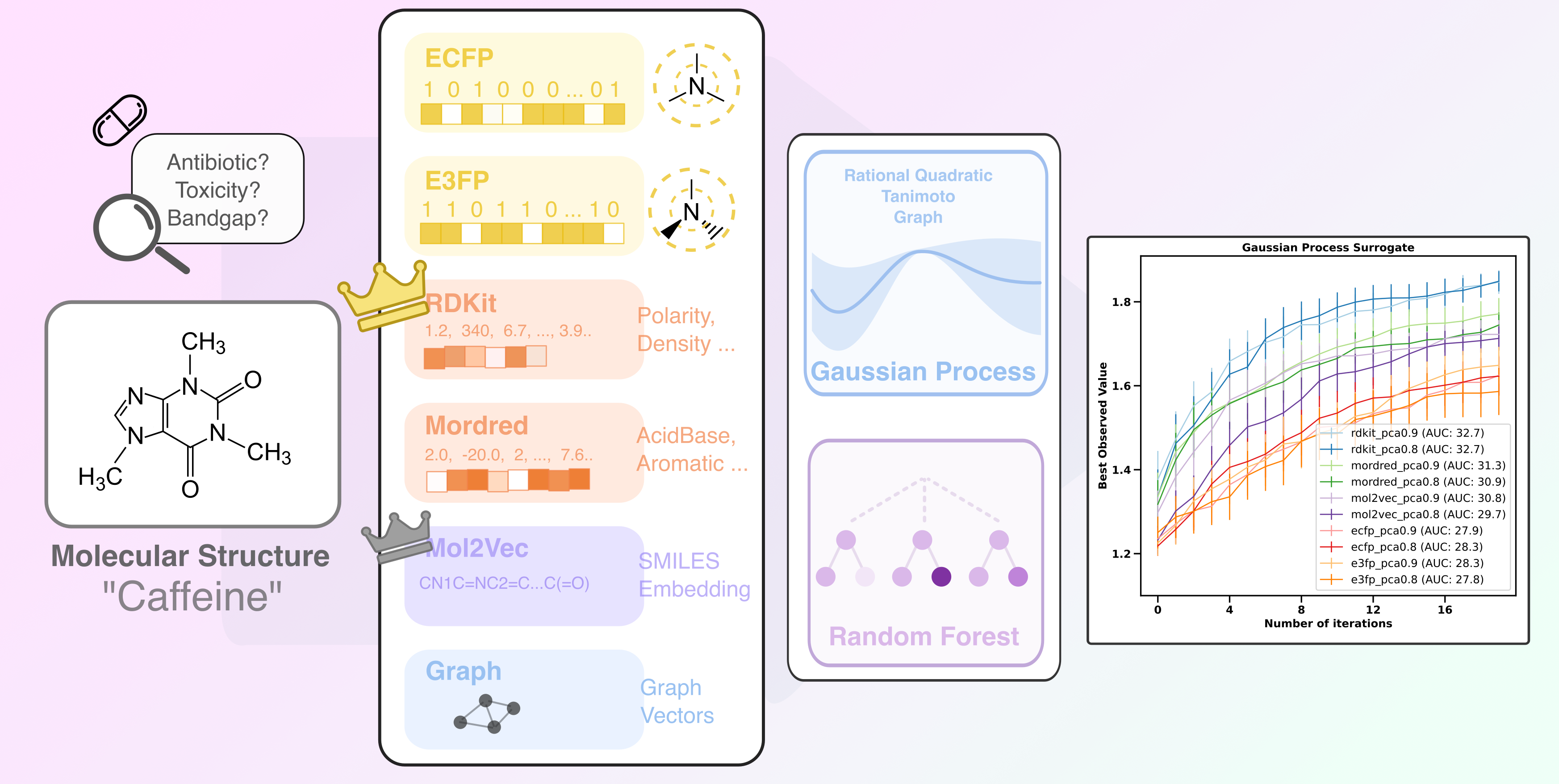
Results
1. Raw Feature Benchmark
mol2vecperformed the best in our Raw Feature benchmark with GP-based surrogates!- The high dimensionality (>1,500) of
mordredmade its raw feature impossible to be incorporated without processing graphrepresentations andgraphkernels were found to be highly resource-demaning in GP-BO, thus not investigatedRF-based surrogatesbrought increased performance withmordredandgraph2vecfeaturizations, but demand significantly more resources to train (due to hyperparameter tuning step in each iter) and exhibit high variability
2. PCAed Feature Benchmark
- With PCA, physicochemcial featurizations such as
rdkitandmordredwon the benchmark. Preserving 90% of variance,rdkitandmordredhad 46 and 50 features, respectively. - Further reduction of features would not further benefit the performance, but will contribute to a shorter runtime.
- PCA is detrimental to latent space featurizations with
mol2vec, we would expect similar obervation withgraph
3. Conclusion and disclaimers
Physicochemical featurizationwith PCA is overall recommended for BO, considering their performance and preservation of chemical information when compared with other representations.- Due to time constraints, our benchmark was on one dataset (lipophilicity - DeepChem). Further exploration are needed for more robust conclusion.
- In our benchmark,
Tanimoto Kernelwas used for bit-string connectivity fingerprint.Rational Quadratic Kernelwas used for physicochemical featurization. The kernel in GP would greatly impact the surrogate’s accuracy and thus need further investigation. But it is beyond the scope (and resource) of the present project.
To reproduce our work
Prerequisites
To replicate the work, the following dependancies are necessary:
- python 3.9
- botorch (framework for BO)
- deepchem (framework for cheminformatics)
- mol2vec (featurization)
- numpy
- scikit-learn
- scikit-optimize
- torch
- gpytorch
To set up the environment, follow the steps:
conda create --name bofeat python=3.9
conda activate bofeat
pip install botorch deepchem numpy scikit-learn scikit-optimize torch gpytorch
pip install git+https://github.com/samoturk/mol2vec
P.S. Installation with conda manager is not recommended as it caused weird incompatibility issue.
P.P.S. Make sure pip is from the newly created bofeat environment. If you’re using a Unix-based OS, you can use which pip to check